Dentate gyrus dataset¶
This tutorial uses raw data from scvelo package. Special thanks go to the scvelo team!
[ ]:
# get the latest version from pypi
# for other installations approaches, see https://dynamo-release.readthedocs.io/en/latest/ten_minutes_to_dynamo.html#how-to-install
!pip install dynamo-release --upgrade --quiet
[1]:
# from IPython.core.display import display, HTML
# display(HTML("<style>.container { width:90% !important; }</style>"))
# %matplotlib inline
import warnings
warnings.filterwarnings('ignore')
import dynamo as dyn
dyn.get_all_dependencies_version()
| package | dynamo-release | umap-learn | anndata | cvxopt | hdbscan | loompy | matplotlib | numba | numpy | pandas | pynndescent | python-igraph | scikit-learn | scipy | seaborn | setuptools | statsmodels | tqdm | trimap | numdifftools | colorcet |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| version | 0.95.2 | 0.4.6 | 0.7.4 | 1.2.3 | 0.8.26 | 3.0.6 | 3.3.0 | 0.51.0 | 1.19.1 | 1.1.1 | 0.4.8 | 0.8.2 | 0.23.2 | 1.5.2 | 0.9.0 | 49.6.0 | 0.11.1 | 4.48.2 | 1.0.12 | 0.9.39 | 2.0.2 |
[2]:
# emulate ggplot2 plotting styple with white background
dyn.configuration.set_figure_params('dynamo', background='white')
[3]:
adata = dyn.sample_data.DentateGyrus_scvelo()
adata.obsm['X_umap_ori'] = adata.obsm['X_umap'].copy()
[4]:
dyn.pl.show_fraction(adata)
dyn.pl.basic_stats(adata)
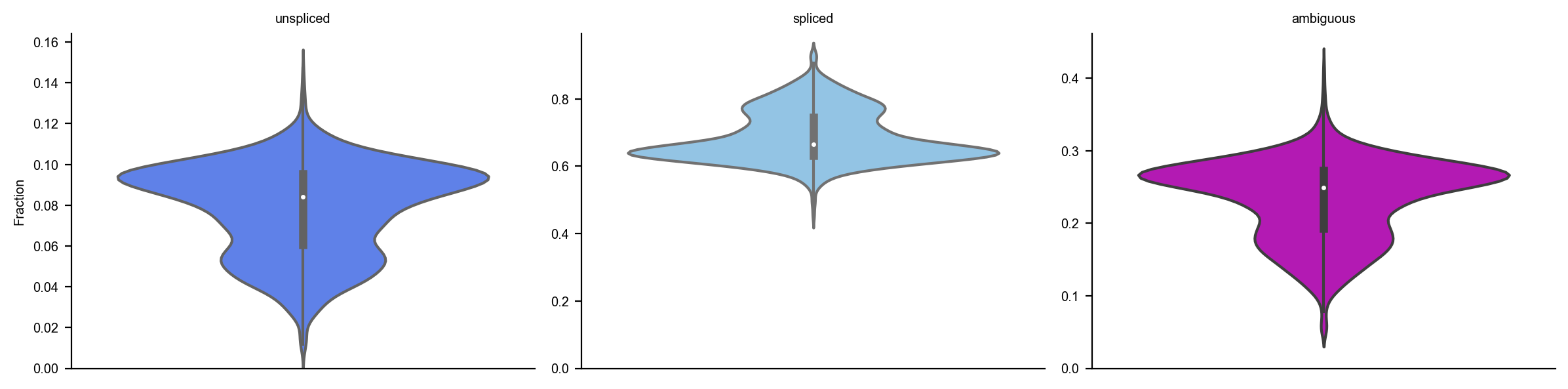
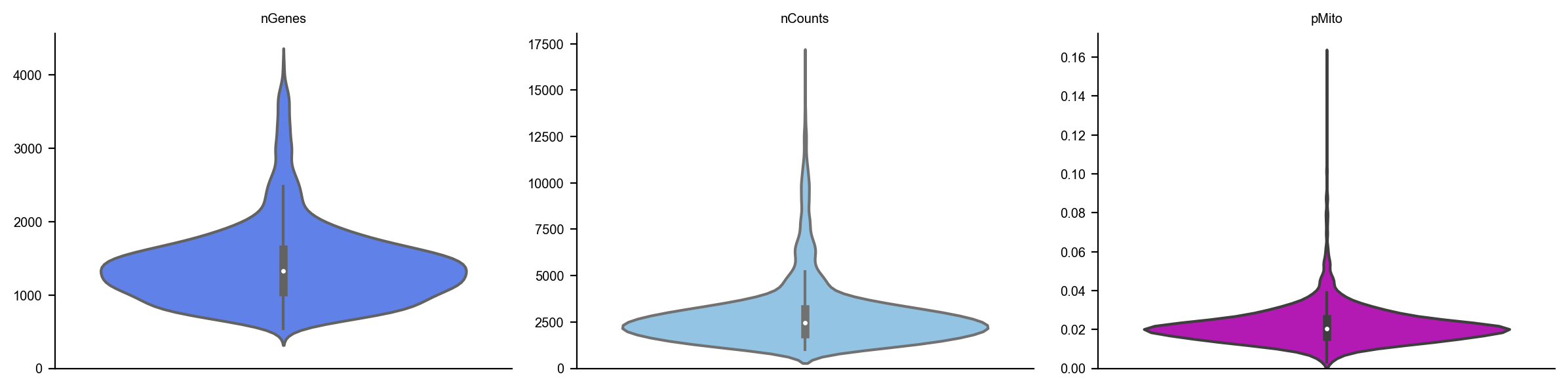
[5]:
dyn.pp.recipe_monocle(adata, n_top_genes=2000, fg_kwargs={'shared_count': 30})
[5]:
AnnData object with n_obs × n_vars = 2930 × 13913
obs: 'clusters', 'age(days)', 'clusters_enlarged', 'nGenes', 'nCounts', 'pMito', 'use_for_pca', 'unspliced_Size_Factor', 'initial_unspliced_cell_size', 'Size_Factor', 'initial_cell_size', 'spliced_Size_Factor', 'initial_spliced_cell_size', 'ntr', 'cell_cycle_phase'
var: 'pass_basic_filter', 'log_m', 'log_cv', 'score', 'use_for_pca', 'ntr'
uns: 'clusters_colors', 'velocyto_SVR', 'pp_norm_method', 'PCs', 'explained_variance_ratio_', 'pca_fit', 'feature_selection'
obsm: 'X_umap', 'X_umap_ori', 'X_pca', 'X', 'cell_cycle_scores'
layers: 'ambiguous', 'spliced', 'unspliced', 'X_unspliced', 'X_spliced'
[6]:
dyn.pl.variance_explained(adata)
dyn.pl.feature_genes(adata)
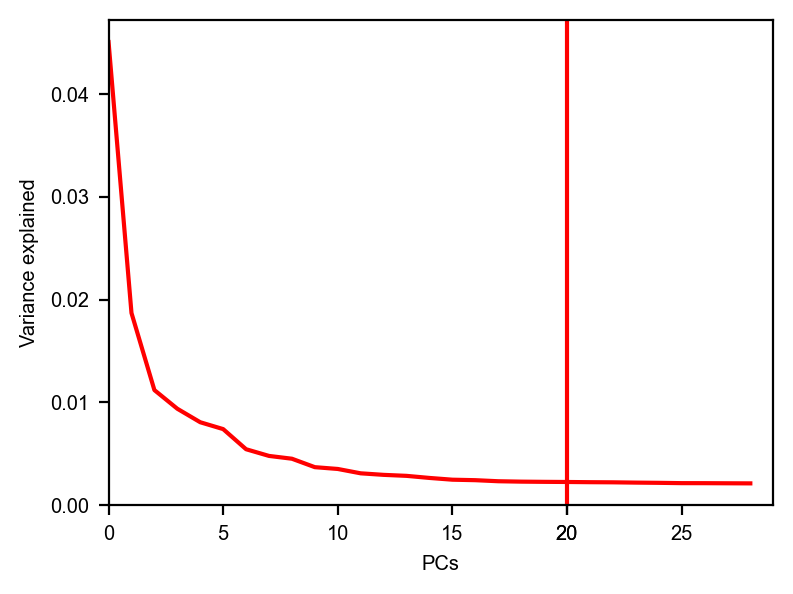
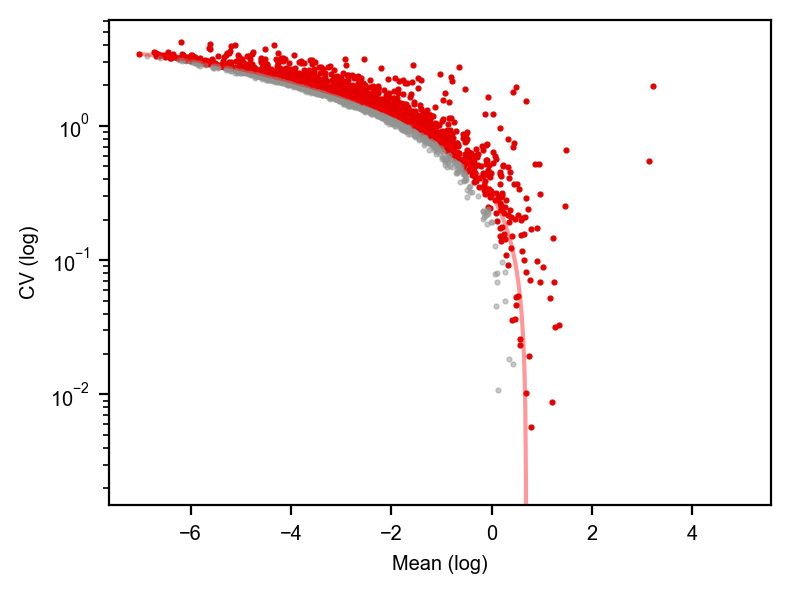
[7]:
dyn.tl.dynamics(adata, model='stochastic', cores=3)
# or dyn.tl.dynamics(adata, model='deterministic')
# or dyn.tl.dynamics(adata, model='stochastic', est_method='negbin')
[7]:
AnnData object with n_obs × n_vars = 2930 × 13913
obs: 'clusters', 'age(days)', 'clusters_enlarged', 'nGenes', 'nCounts', 'pMito', 'use_for_pca', 'unspliced_Size_Factor', 'initial_unspliced_cell_size', 'Size_Factor', 'initial_cell_size', 'spliced_Size_Factor', 'initial_spliced_cell_size', 'ntr', 'cell_cycle_phase'
var: 'pass_basic_filter', 'log_m', 'log_cv', 'score', 'use_for_pca', 'ntr', 'beta', 'gamma', 'half_life', 'alpha_b', 'alpha_r2', 'gamma_b', 'gamma_r2', 'gamma_logLL', 'delta_b', 'delta_r2', 'uu0', 'ul0', 'su0', 'sl0', 'U0', 'S0', 'total0', 'use_for_dynamics'
uns: 'clusters_colors', 'velocyto_SVR', 'pp_norm_method', 'PCs', 'explained_variance_ratio_', 'pca_fit', 'feature_selection', 'dynamics'
obsm: 'X_umap', 'X_umap_ori', 'X_pca', 'X', 'cell_cycle_scores'
layers: 'ambiguous', 'spliced', 'unspliced', 'X_unspliced', 'X_spliced', 'M_u', 'M_uu', 'M_s', 'M_us', 'M_ss', 'velocity_S'
obsp: 'moments_con'
[8]:
# enforce recalculating the umap embedding. By default dynamo will avoid recalculation if a reduced dimension space exists.
dyn.tl.reduceDimension(adata, enforce=True)
[8]:
AnnData object with n_obs × n_vars = 2930 × 13913
obs: 'clusters', 'age(days)', 'clusters_enlarged', 'nGenes', 'nCounts', 'pMito', 'use_for_pca', 'unspliced_Size_Factor', 'initial_unspliced_cell_size', 'Size_Factor', 'initial_cell_size', 'spliced_Size_Factor', 'initial_spliced_cell_size', 'ntr', 'cell_cycle_phase'
var: 'pass_basic_filter', 'log_m', 'log_cv', 'score', 'use_for_pca', 'ntr', 'beta', 'gamma', 'half_life', 'alpha_b', 'alpha_r2', 'gamma_b', 'gamma_r2', 'gamma_logLL', 'delta_b', 'delta_r2', 'uu0', 'ul0', 'su0', 'sl0', 'U0', 'S0', 'total0', 'use_for_dynamics'
uns: 'clusters_colors', 'velocyto_SVR', 'pp_norm_method', 'PCs', 'explained_variance_ratio_', 'pca_fit', 'feature_selection', 'dynamics', 'neighbors', 'umap_fit'
obsm: 'X_umap', 'X_umap_ori', 'X_pca', 'X', 'cell_cycle_scores'
layers: 'ambiguous', 'spliced', 'unspliced', 'X_unspliced', 'X_spliced', 'M_u', 'M_uu', 'M_s', 'M_us', 'M_ss', 'velocity_S'
obsp: 'moments_con', 'connectivities', 'distances'
[9]:
dyn.tl.cell_velocities(adata)
calculating transition matrix via pearson kernel with sqrt transform.: 100%|██████████| 2930/2930 [00:05<00:00, 498.05it/s]
projecting velocity vector to low dimensional embedding...: 100%|██████████| 2930/2930 [00:00<00:00, 3557.37it/s]
[9]:
AnnData object with n_obs × n_vars = 2930 × 13913
obs: 'clusters', 'age(days)', 'clusters_enlarged', 'nGenes', 'nCounts', 'pMito', 'use_for_pca', 'unspliced_Size_Factor', 'initial_unspliced_cell_size', 'Size_Factor', 'initial_cell_size', 'spliced_Size_Factor', 'initial_spliced_cell_size', 'ntr', 'cell_cycle_phase'
var: 'pass_basic_filter', 'log_m', 'log_cv', 'score', 'use_for_pca', 'ntr', 'beta', 'gamma', 'half_life', 'alpha_b', 'alpha_r2', 'gamma_b', 'gamma_r2', 'gamma_logLL', 'delta_b', 'delta_r2', 'uu0', 'ul0', 'su0', 'sl0', 'U0', 'S0', 'total0', 'use_for_dynamics', 'use_for_transition'
uns: 'clusters_colors', 'velocyto_SVR', 'pp_norm_method', 'PCs', 'explained_variance_ratio_', 'pca_fit', 'feature_selection', 'dynamics', 'neighbors', 'umap_fit', 'grid_velocity_umap'
obsm: 'X_umap', 'X_umap_ori', 'X_pca', 'X', 'cell_cycle_scores', 'velocity_umap'
layers: 'ambiguous', 'spliced', 'unspliced', 'X_unspliced', 'X_spliced', 'M_u', 'M_uu', 'M_s', 'M_us', 'M_ss', 'velocity_S'
obsp: 'moments_con', 'connectivities', 'distances', 'pearson_transition_matrix'
[10]:
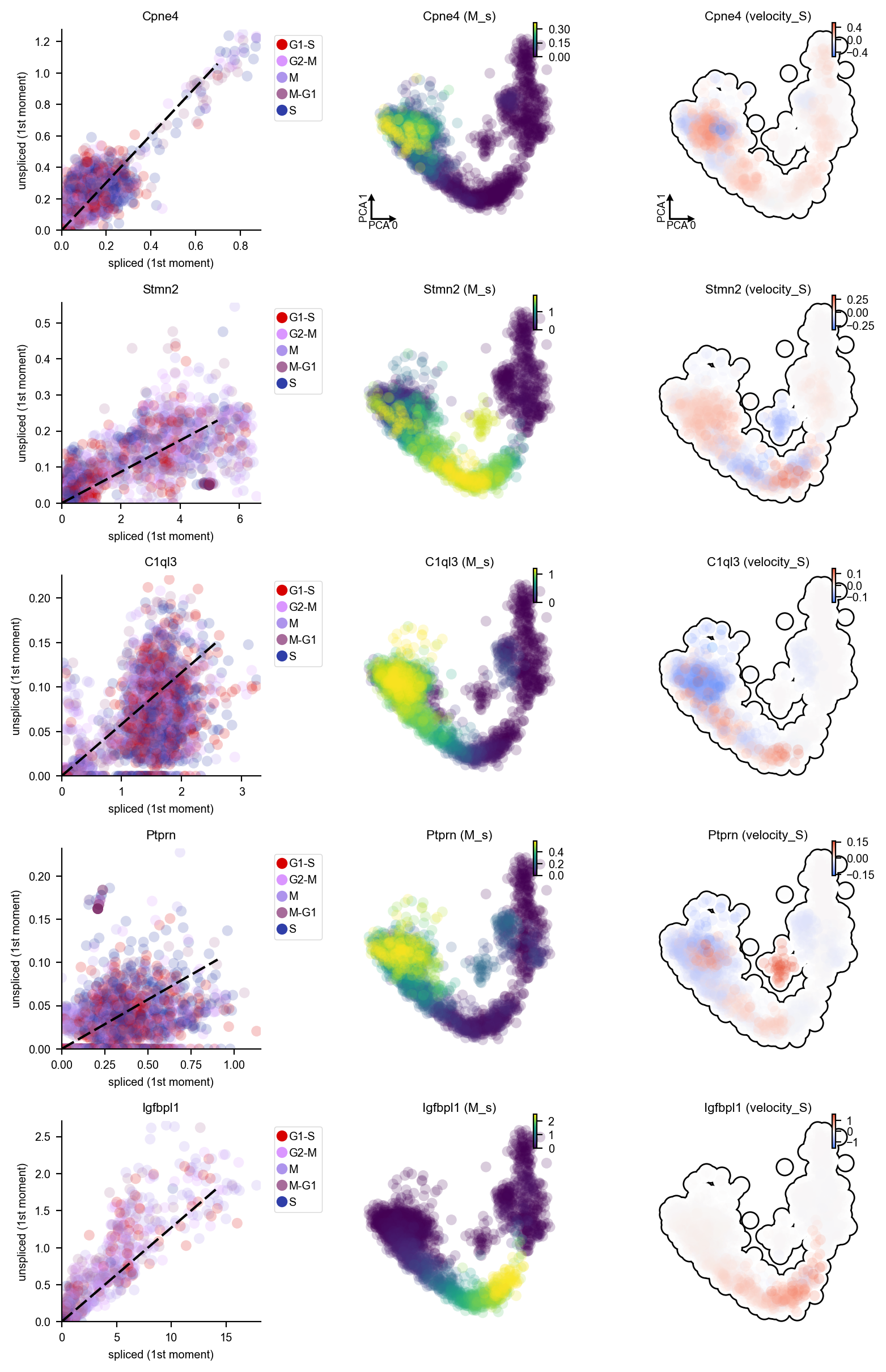
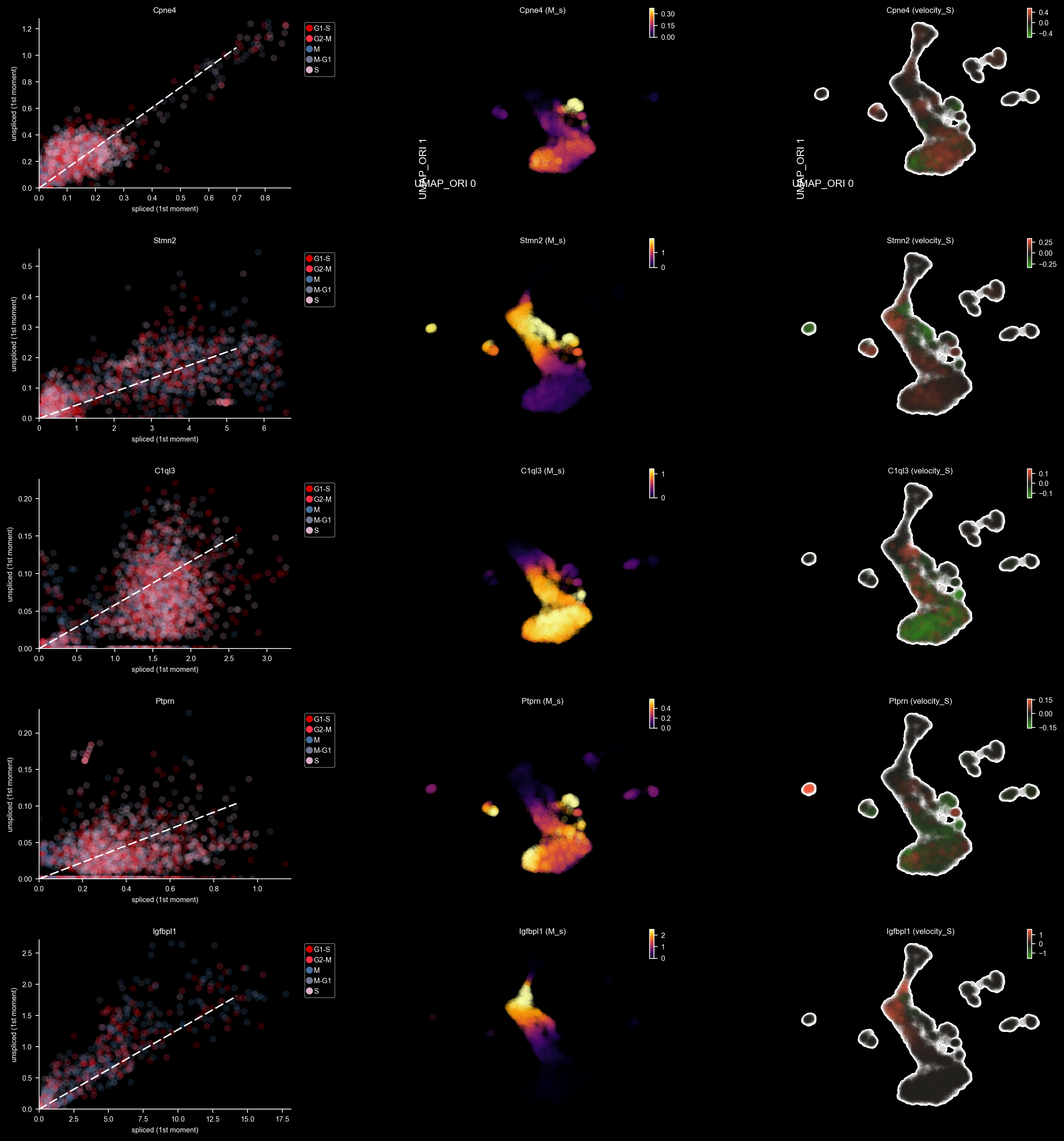
DentateGyrus_genes = ["Tnc", "Gfap", "Tac2", "Pdgfra", "Igfbpl1", 'Ptprn', "Sema3c", "Neurod6", "Stmn2", "Sema5a", "C1ql3", "Cpne4", "Cck"]
dyn.pl.phase_portraits(adata, genes=DentateGyrus_genes, ncols=3, figsize=(3, 3), basis='pca', show_quiver=False)
[11]:
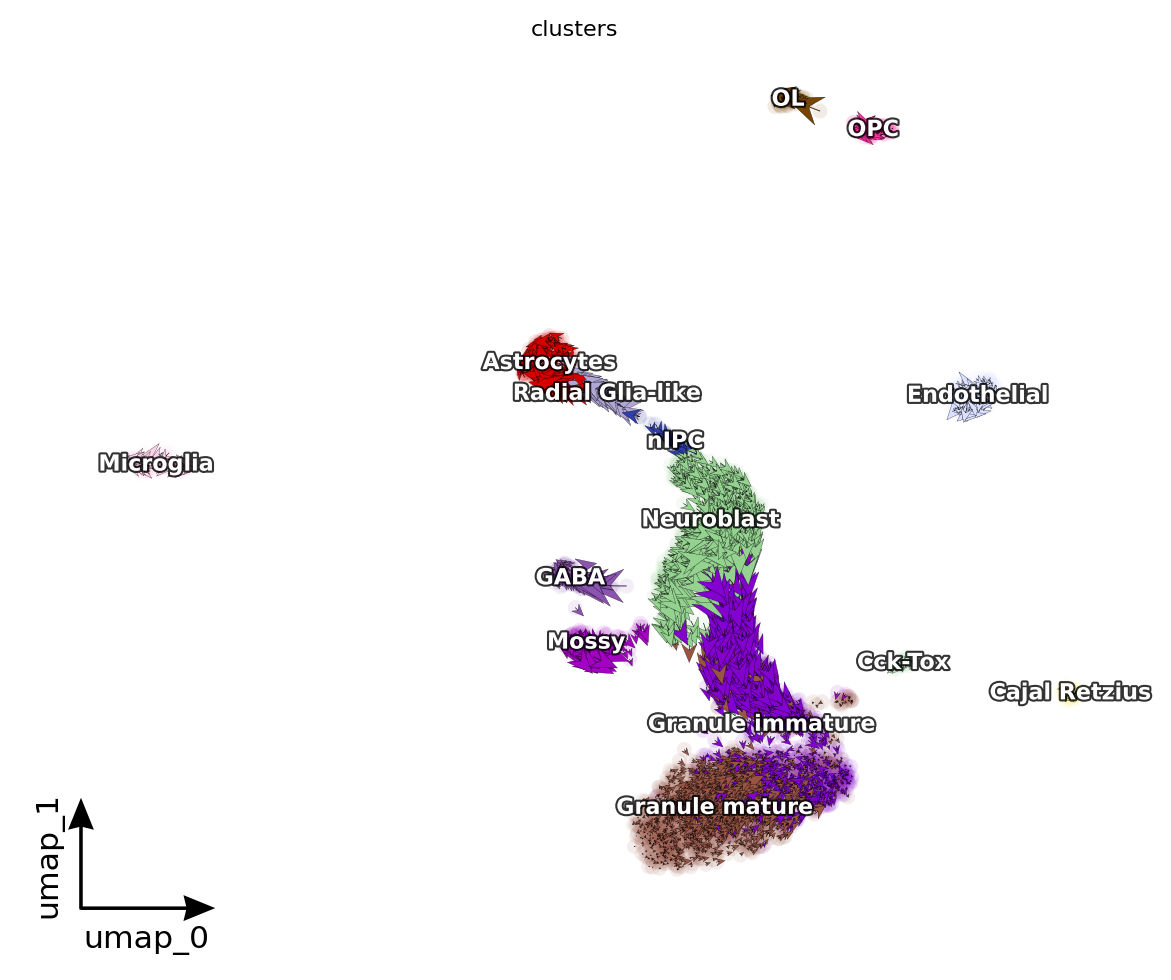
dyn.pl.cell_wise_vectors(adata, color=['clusters'], quiver_size=6, quiver_length=6, figsize=(6, 5), pointsize=0.1) # ['GRIA3', 'LINC00982', 'AFF2']
<Figure size 600x500 with 0 Axes>
[12]:
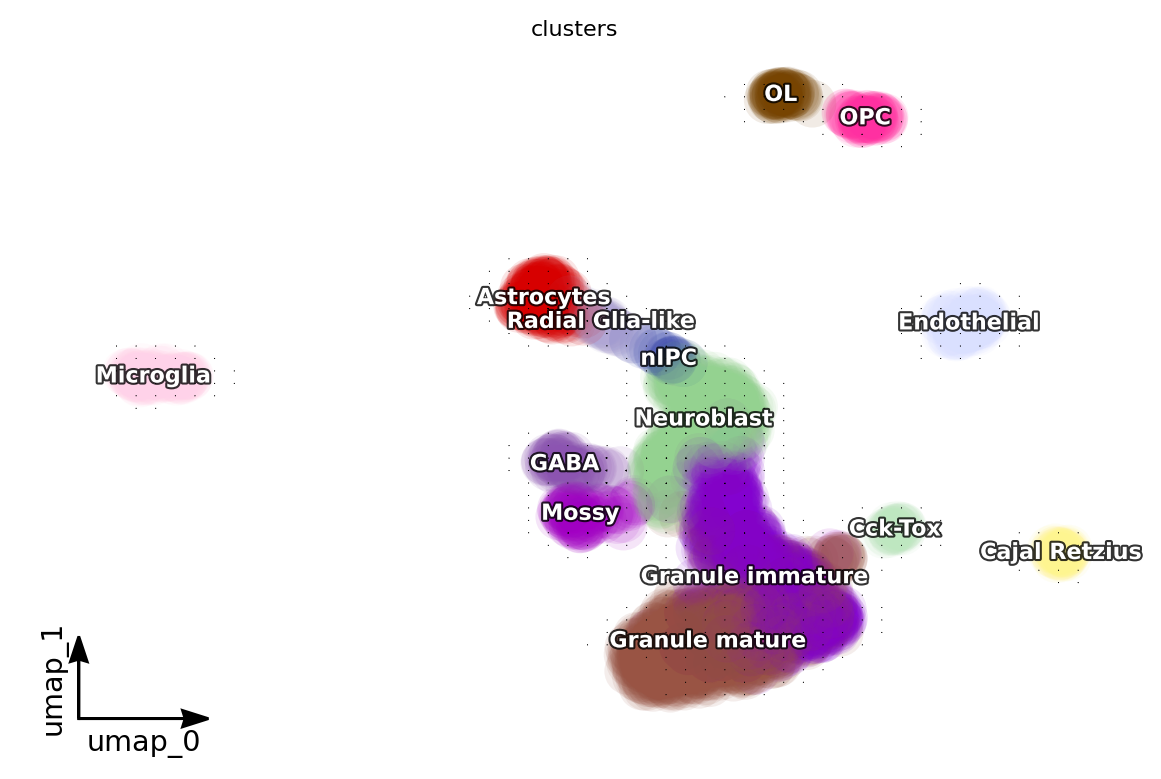
dyn.pl.grid_vectors(adata, color=['clusters'], method='gaussian')
<Figure size 600x400 with 0 Axes>
[13]:
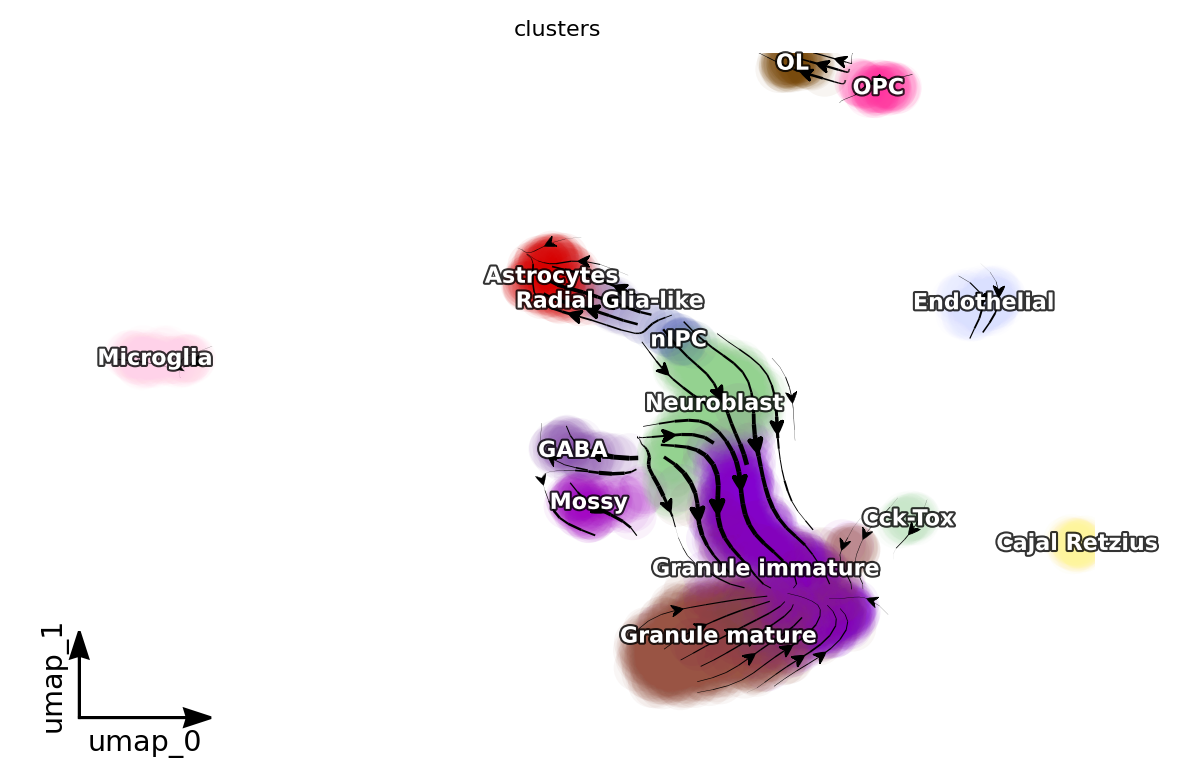
dyn.pl.streamline_plot(adata, color=['clusters'], basis='umap', density=1, background='white',s_kwargs_dict={"alpha": 0.05})
<Figure size 600x400 with 0 Axes>
[14]:
dyn.pl.streamline_plot(adata, color=['clusters'], basis='umap_ori', density=1, background='white',s_kwargs_dict={"alpha": 0.05})
projecting velocity vector to low dimensional embedding...: 27%|██▋ | 784/2930 [00:00<00:00, 3801.14it/s]
Using existing pearson_transition_matrix found in .obsp.
projecting velocity vector to low dimensional embedding...: 100%|██████████| 2930/2930 [00:00<00:00, 4048.10it/s]
<Figure size 600x400 with 0 Axes>
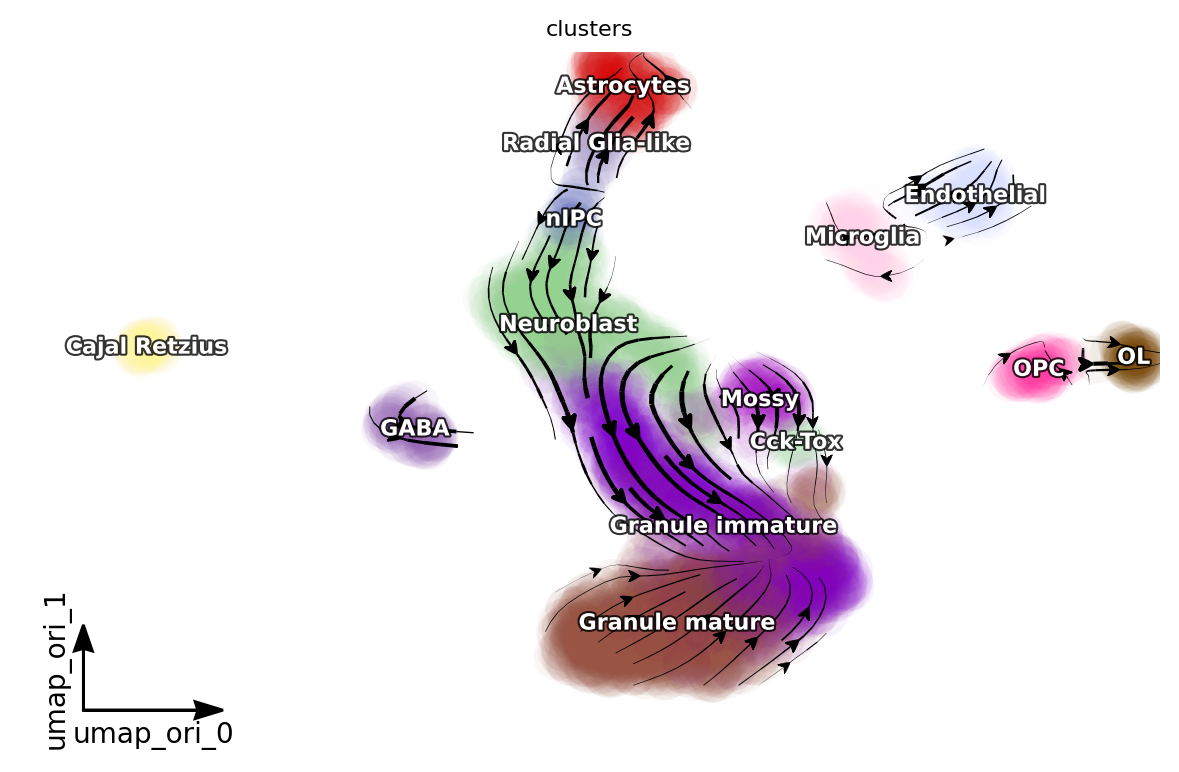
[15]:
# you can set `verbose = 1/2/3` to obtain different levels of running information of vector field reconstruction
dyn.vf.VectorField(adata, basis='umap_ori', dims=[0, 1])
[16]:
dyn.pl.topography(adata, color=['clusters', 'Tnc'], basis='umap_ori', ncols=2)
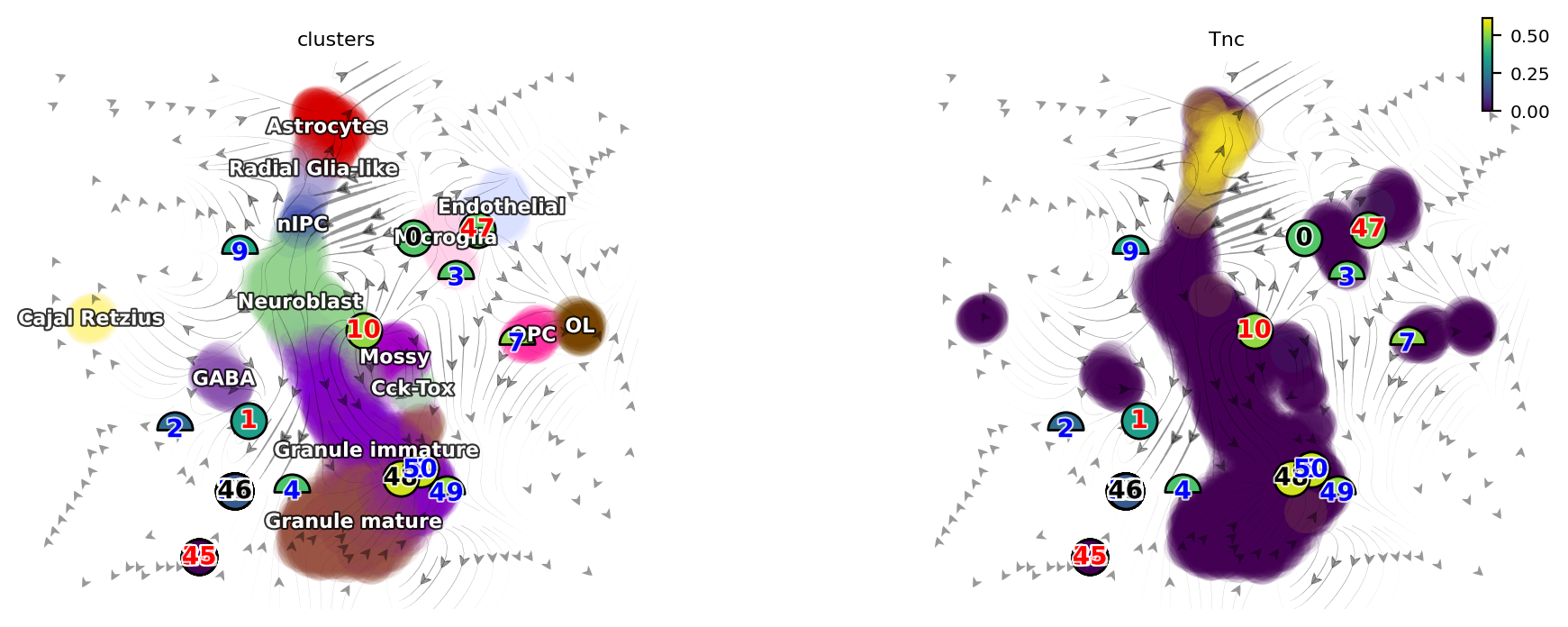
[17]:
dyn.vf.VectorField(adata, basis='umap_ori', dims=[0, 1], pot_curl_div=True)
[18]:
dyn.ext.ddhodge(adata, basis='umap_ori')
Constructing diffusion graph from reconstructed vector field: 2930it [01:57, 24.83it/s]
[19]:
adata
[19]:
AnnData object with n_obs × n_vars = 2930 × 13913
obs: 'clusters', 'age(days)', 'clusters_enlarged', 'nGenes', 'nCounts', 'pMito', 'use_for_pca', 'unspliced_Size_Factor', 'initial_unspliced_cell_size', 'Size_Factor', 'initial_cell_size', 'spliced_Size_Factor', 'initial_spliced_cell_size', 'ntr', 'cell_cycle_phase', 'umap_ori_ddhodge_div', 'umap_ori_ddhodge_potential'
var: 'pass_basic_filter', 'log_m', 'log_cv', 'score', 'use_for_pca', 'ntr', 'beta', 'gamma', 'half_life', 'alpha_b', 'alpha_r2', 'gamma_b', 'gamma_r2', 'gamma_logLL', 'delta_b', 'delta_r2', 'uu0', 'ul0', 'su0', 'sl0', 'U0', 'S0', 'total0', 'use_for_dynamics', 'use_for_transition'
uns: 'clusters_colors', 'velocyto_SVR', 'pp_norm_method', 'PCs', 'explained_variance_ratio_', 'pca_fit', 'feature_selection', 'dynamics', 'neighbors', 'umap_fit', 'grid_velocity_umap', 'grid_velocity_umap_ori', 'VecFld_umap_ori', 'VecFld'
obsm: 'X_umap', 'X_umap_ori', 'X_pca', 'X', 'cell_cycle_scores', 'velocity_umap', 'velocity_umap_ori', 'velocity_umap_ori_SparseVFC', 'X_umap_ori_SparseVFC'
layers: 'ambiguous', 'spliced', 'unspliced', 'X_unspliced', 'X_spliced', 'M_u', 'M_uu', 'M_s', 'M_us', 'M_ss', 'velocity_S'
obsp: 'moments_con', 'connectivities', 'distances', 'pearson_transition_matrix', 'umap_ori_ddhodge'
[20]:
dyn.pl.topography(adata, color=['clusters', 'umap_ori_ddhodge_div', 'umap_ori_ddhodge_potential'],
basis='umap_ori', ncols=3, frontier=True)
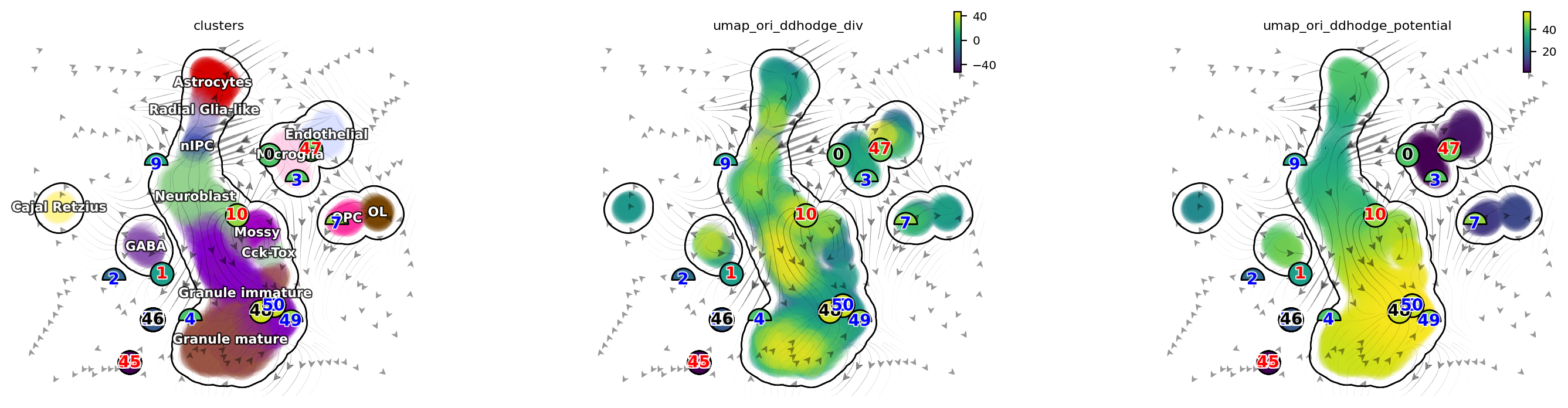
[21]:
# emulate ggplot2 plotting styple with black background
dyn.configuration.set_figure_params('dynamo', background='black')
[22]:
dyn.pl.phase_portraits(adata, genes=DentateGyrus_genes, ncols=3, figsize=(6, 4), basis='umap_ori', show_quiver=False)
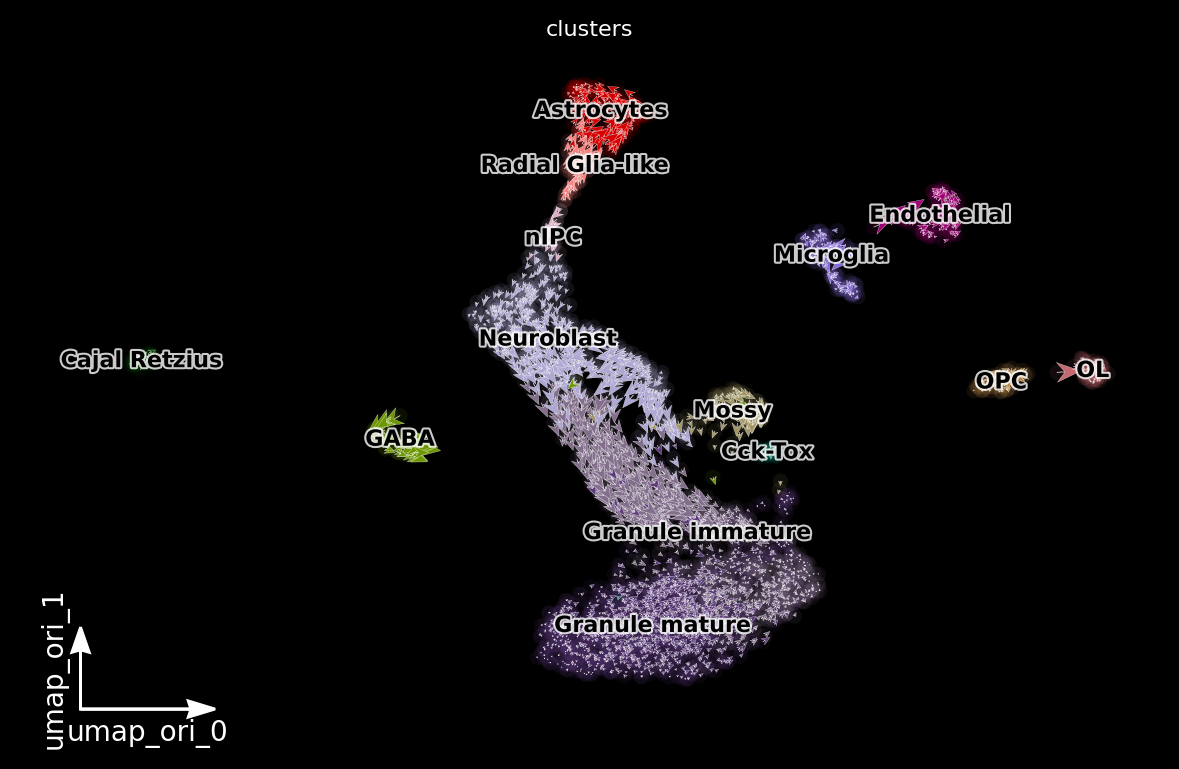
dyn.pl.cell_wise_vectors(adata, color=['clusters'], basis='umap_ori', pointsize=0.1, quiver_size=4, quiver_length=4, background='black') # ['GRIA3', 'LINC00982', 'AFF2']
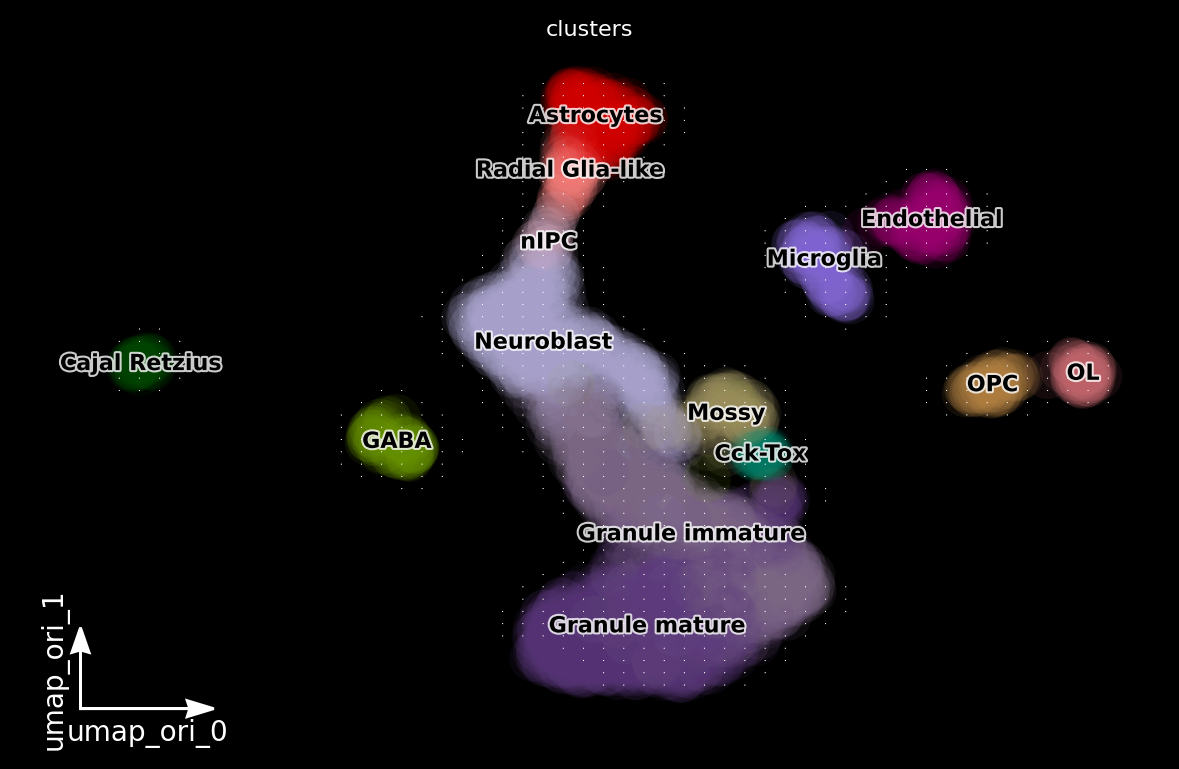
dyn.pl.grid_vectors(adata, color=['clusters'], basis='umap_ori', method='gaussian', background='black')
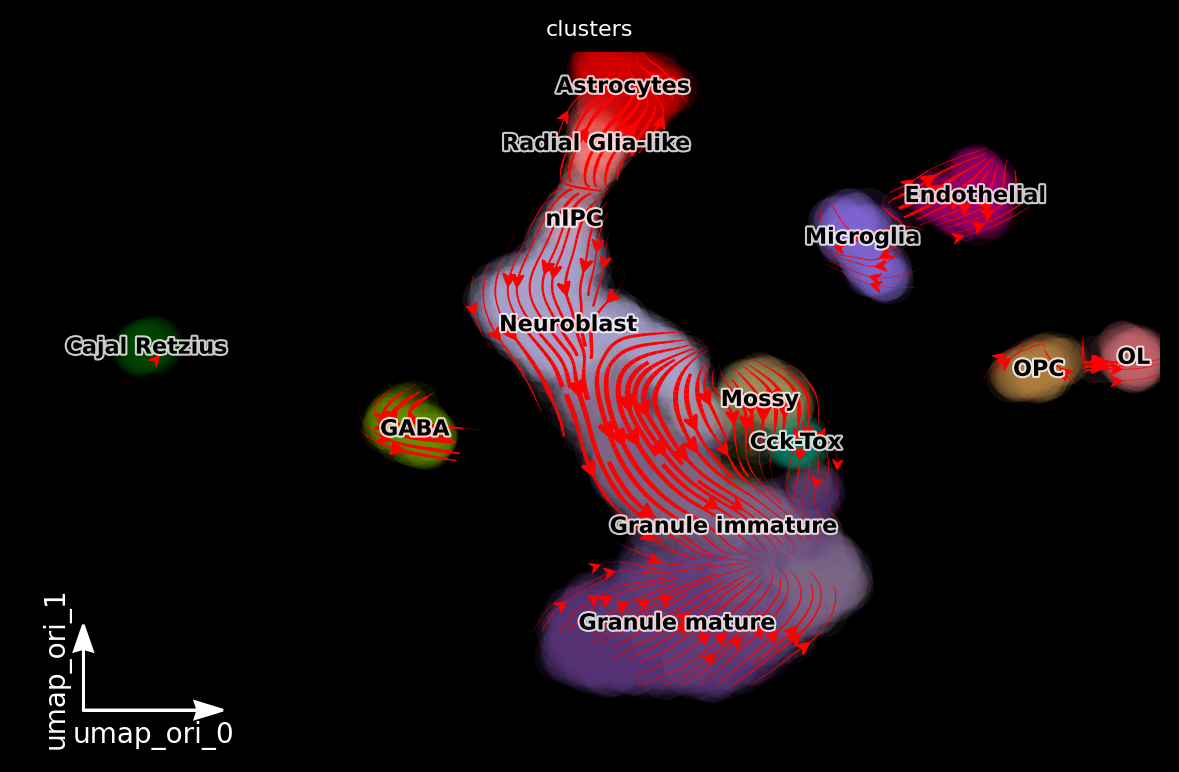
dyn.pl.streamline_plot(adata, color=['clusters'], basis='umap_ori', density=2, background='black')
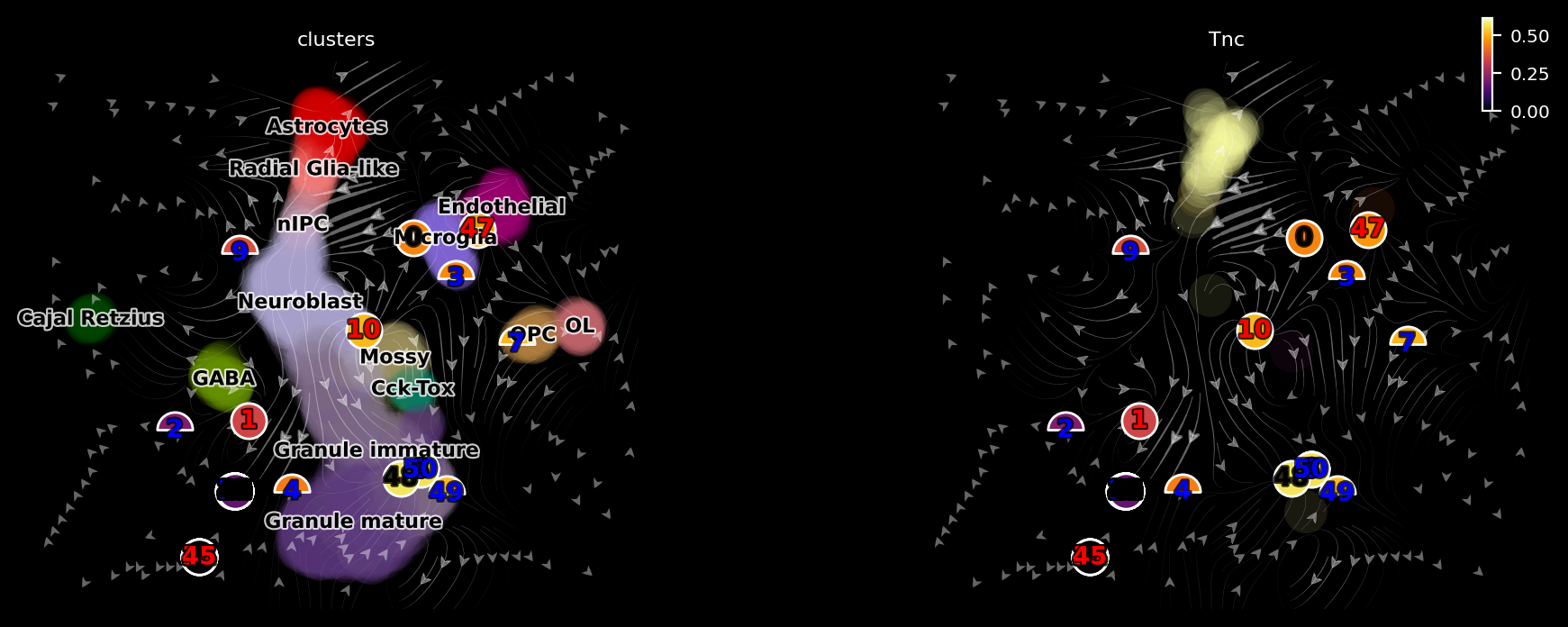
dyn.pl.topography(adata, color=['clusters', 'Tnc'], basis='umap_ori', ncols=2, background='black')
<Figure size 600x400 with 0 Axes>
<Figure size 600x400 with 0 Axes>
<Figure size 600x400 with 0 Axes>
[23]:
progenitor = adata.obs_names[adata.obs.clusters.isin(['nIPC', 'Neuroblast'])]
len(progenitor)
[23]:
436
[24]:
dyn.pd.fate(adata, basis='umap_ori', init_cells=progenitor, interpolation_num=100, direction='forward',
inverse_transform=False, average=False, cores=3)
[24]:
AnnData object with n_obs × n_vars = 2930 × 13913
obs: 'clusters', 'age(days)', 'clusters_enlarged', 'nGenes', 'nCounts', 'pMito', 'use_for_pca', 'unspliced_Size_Factor', 'initial_unspliced_cell_size', 'Size_Factor', 'initial_cell_size', 'spliced_Size_Factor', 'initial_spliced_cell_size', 'ntr', 'cell_cycle_phase', 'umap_ori_ddhodge_div', 'umap_ori_ddhodge_potential'
var: 'pass_basic_filter', 'log_m', 'log_cv', 'score', 'use_for_pca', 'ntr', 'beta', 'gamma', 'half_life', 'alpha_b', 'alpha_r2', 'gamma_b', 'gamma_r2', 'gamma_logLL', 'delta_b', 'delta_r2', 'uu0', 'ul0', 'su0', 'sl0', 'U0', 'S0', 'total0', 'use_for_dynamics', 'use_for_transition'
uns: 'clusters_colors', 'velocyto_SVR', 'pp_norm_method', 'PCs', 'explained_variance_ratio_', 'pca_fit', 'feature_selection', 'dynamics', 'neighbors', 'umap_fit', 'grid_velocity_umap', 'grid_velocity_umap_ori', 'VecFld_umap_ori', 'VecFld', 'fate_umap_ori'
obsm: 'X_umap', 'X_umap_ori', 'X_pca', 'X', 'cell_cycle_scores', 'velocity_umap', 'velocity_umap_ori', 'velocity_umap_ori_SparseVFC', 'X_umap_ori_SparseVFC'
layers: 'ambiguous', 'spliced', 'unspliced', 'X_unspliced', 'X_spliced', 'M_u', 'M_uu', 'M_s', 'M_us', 'M_ss', 'velocity_S'
obsp: 'moments_con', 'connectivities', 'distances', 'pearson_transition_matrix', 'umap_ori_ddhodge'
[25]:
%%capture
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
ax = dyn.pl.topography(adata, color='clusters', ax=ax, save_show_or_return='return', basis='umap_ori')
[26]:
%%capture
instance = dyn.mv.StreamFuncAnim(adata=adata, ax=ax, basis='umap_ori', color='clusters')
[27]:
from matplotlib import animation
import numpy as np
anim = animation.FuncAnimation(instance.fig, instance.update, init_func=instance.init_background,
frames=np.arange(100), interval=100, blit=True)
from IPython.core.display import display, HTML
HTML(anim.to_jshtml()) # embedding to jupyter notebook.
[27]: